About
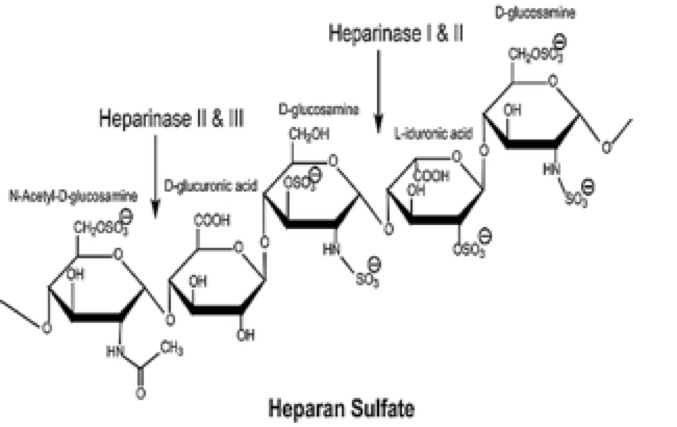
Pedobacter heparinus is a non-pathogenic, free living, gram-negative rod shaped soil bacterium first isolated by Payza and Korn (1956). This bacterium was unusual since it could grow on heparin or sulfated acidic mucoheteropolysaccharides from various animal tissues as the sole source of carbon, nitrogen and energy. Five glycosaminoglycan-degrading enzymes have been purified from P. heparinus. These include heparinases I, II and III (see figure below) and chondroitinases AC and B (Yang et al. 1985). Eventually the genes for these enzymes were cloned to produce large quantities of the pure enzymes for commercial and research applications. For example, the heparinase enzymes were used to study structure of heparin, a drug widely used as an anticoagulant.

In spite of the discovery of these unique polysaccharide degrading enzymes, very little is known about the P. heparinus as a free living environmental bacterium. There are now 50 species in the Pedobacter genus and heparinus is the type species for the genus. However, as of 2015, there have only been about 20 published papers on this type species and mostly focused on enzyme purification.
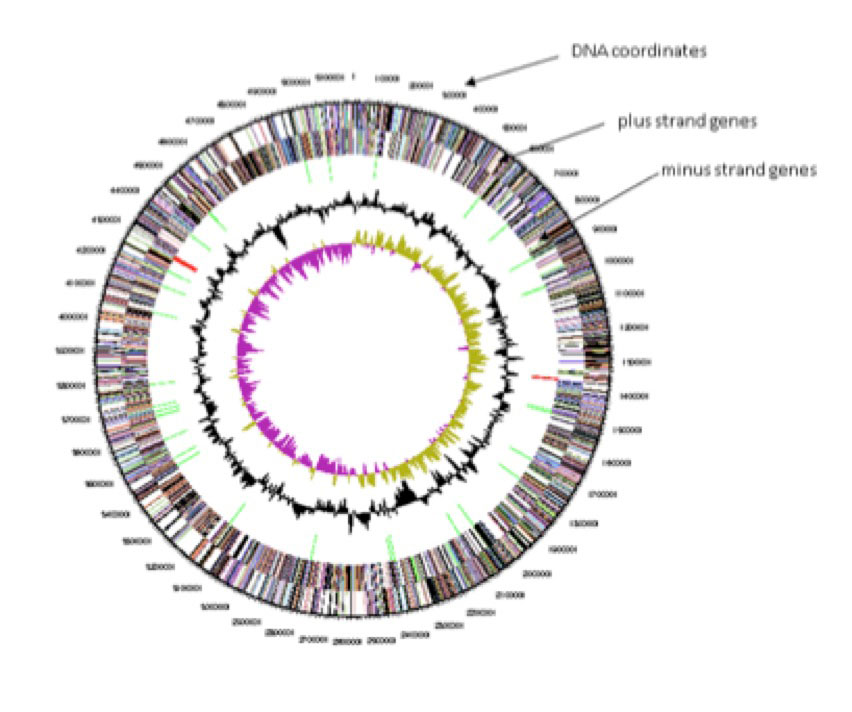
In 2009, Han and associates at the DOE Joint Genome Institute (JGI) published the complete genome sequence of P. heparinus. The organism was selected for sequencing as a type species organism for the genus Pedobacter and was part of the Genomic Encyclopedia of Bacteria and Archaea (GEBA) project. The circular genome is 5,167,383 bp long. Using a program called Prodigal, the DNA sequence was predicted to contain 4,341 genes, of which 4,287 were protein coding and 54 RNA coding.
In 2010, Geneva College was selected to participate in the Interpret a Genome in Education sponsored by the JGI. A major goal of this program was to “annotate” the genomes of newly sequenced genomes type species from a genus not well studied. Annotation is a type of manual curation in which of the gene in the genome is further studied to confirm the predictions by Prodigal. The gene curation was to be done by “massively parallel undergraduates” thus creating a research experience for many students and allowing for high throughput. Biology professor Dr. David Essig and Chemistry professor, Dr. Rodney Austin attended the January 2010 Bioinformatics workshop held at the JGI in Walnut Creek, CA. Shortly thereafter, workshop participants were encouraged to “adopt” the genome of an organism. Drs. Essig and Austin chose P. heparinus given that it was a relatively uncharacterized type of species (as described above), but also because it could be easily cultured at room temperature (a mesophile) in the presence of oxygen.

Since 2010, Geneva College Biology and Chemistry majors in several courses and independent studies have improved the annotation of over 300 genes and carried out experiments to test hypotheses from bioinformatic data. Some findings of interest include
- Besides heparinus, P. heparinus genome encodes genes for many other polysaccharidase enzymes. These include, but are not limited to, celluases and pectinases. These types of genes are found in prototypical “polysaccharide utilization loci” (PUL). A PUL contains a cluster of adjacent genes which includes the polysaccharidase to cleave the polysaccharide into oliogsaccharides (GH family), a gene whose product encodes an extracellular sensor for that polysaccharide (SusD like), a gene for an outer membrane transporter for the polyssacharide (SusC like) and a variety of other genes which code for GH family enzymes which cleave the oliogsaccahrides into mono or disaccharides. The genome of P. heparinus and related species possess the greatest number of PUL’s in comparison to other bacterial species.
- The presence of polysaccharidases such as cellulases and pectinases in P. heparinus raised important questions about niche and habitat. This bacterium could be a decomposer in bulk soil or associated with plant roots (rhizospheric) or within plant roots (endophytic). Based on bioinformatic pathway analysis of the P. heparinus genome, there are genes coding for enzymes involved in plant hormone biosynthesis. This type of evidence points to a plant associated niche.
- Studies by our students have confirmed that P. heparinus is capable of rapid colony spreading on low percent agar in minimal growth medium. Students have also annotated genes for a complete set of gld proteins required for a unique type of surface crawling, which permits the colony spreading.
- Genome annotation studies by our students have shown the presence of genes encoding anti-microbial proteins and peptides. These have included novel lantibiotics, bacterocins, toxin-antitoxins and RHS YPD repeat proteins. Genes for such proteins are likely used in interspecies competition and may also have value as future therapeutic agents.
- A Biology major asked what would happen if P. heparinus were cultured in light instead of dark? Results showed that exposure of P. heparinus cultures promoted yellow pigment production compared to the dark. An operon encoding the enzymes for xeaxanthin has been annotated. The operon also contains a transcription factor as the first gene, which encodes a repressor protein. Studies are underway to understand the light regulation of this operon.
Reference List:
Han, C. and 33 other authors. (2009) Complete genome sequence of Pedobacter heparinus type strain (HIM 762-3T). Standards in Genomic Science 1: 54-62.
Payza, A.N. and Korn, E.D. (1956) Bacterial degradation of heparin. Nature 177: 88-89.
Yang V.C., Linhardt., R. J., Bernstein, H., Cooney, C. L. and Langer. R. (1985). Purification and characterization of heparinase from Flavobacterium heparinium. J Biol Chem 260: 1849 – 1857.